2019: Core architecture of a bacterial type II secretion system
Bacterial type II secretion systems (T2SSs) translocate virulence factors, toxins and enzymes across the cell outer membrane. They constitute part of an ancient type IV filament superfamily that may have been present within the last universal common ancestor (LUCA). Mechanistic principles that underlie T2SS function have implication for other closely related systems such as the type IV and tight adherence pilus systems.
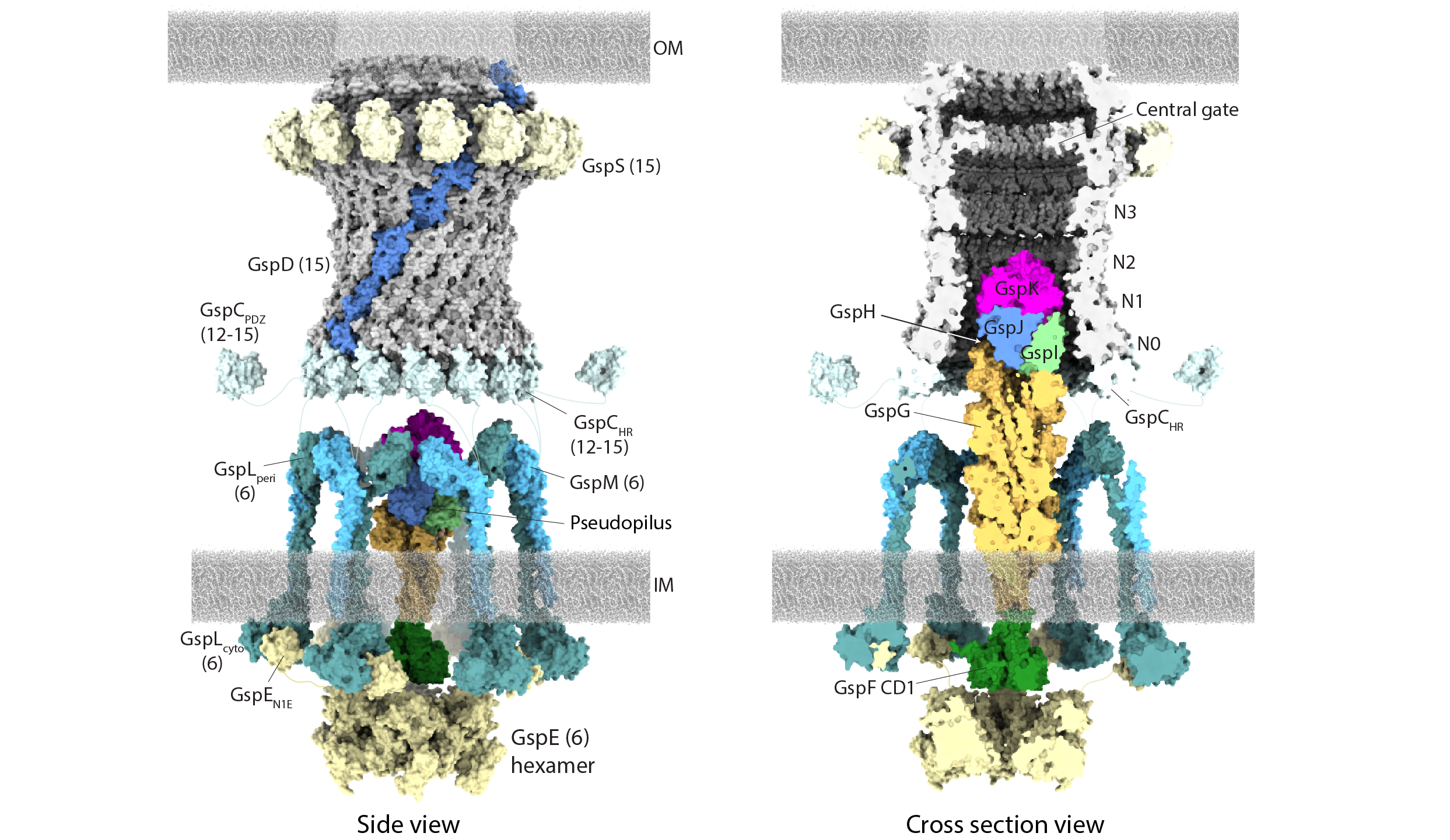
In this project, we use negative stain and cryo-electron microscopy to reveal the core architecture of an assembled T2SS from the pathogen Klebsiella pneumoniae. We show that 7 proteins form a ~2.4 MDa complex that spans the cell envelope. As shown below, the outer membrane complex includes the secretin PulD, with all domains modelled, and the pilotin PulS.
Cryo-EM structure of the T2SS outer membrane complex, which includes the PulD secretin (light blue) and PulS pilotin (cyan) with C15 symmetry. PulC HR domain (orange) is positioned at the base of the secretin.
We also glimpsed the inner membrane assembly platform components PulC, PulE, PulL, PulM and PulN for the first time and probed their stoichiometry. The PulE ATPase, PulL and PulM combine to form a flexible hexameric hub. Symmetry mismatch between the outer membrane complex and assembly platform is overcome by PulC linkers spanning the periplasm, with PulC HR domains binding independently at the secretin base. Overall, our results show that the T2SS has a highly dynamic modular architecture, with implication for pseudo-pilus assembly and substrate loading.
(Left) Negative stain class averages of the PulCDELMNS complex showing the first glimpse of the assembled inner membrane platform. (Right) Stoichiometric model of an assembled T2SS core apparatus. Inner membrane platform components comprise a hexameric hub. PulC connects the inner membrane platform to the outer membrane complex by binding to the secretin base. Substrate may be loaded through the PulC cage in positions where PulC is absent.
2020: The structure and mechanism of the bacterial type II secretion system (Review)
The type II secretion system (T2SS) is a multi-protein complex used by many bacteria to move substrates across their cell membrane. Substrates released into the environment serve as local and long-range effectors that promote nutrient acquisition, biofilm formation and pathogenicity. In both animals and plants, the T2SS is increasingly recognised as a key driver of virulence. The T2SS spans the bacterial cell envelope and extrudes substrates through an outer membrane secretin channel using a pseudopilus. An inner membrane assembly platform and a cytoplasmic motor controls pseudopilus assembly. Advances in cryo-electron microscopy are enabling increasingly elaborate sub-complexes to be resolved (for example check out our paper Chernyatina and Low, Nature Comms, 2018!). However, key questions remain regarding mechanism of pseudopilus extension and retraction, and how this is coupled to the choreography of substrate moving through the secretion system. These questions, as well as the broader structure and mechanism of the T2SS, are discussed in our review.
A model of the bacterial Type II secretion system. Substrates include the cholera and LT toxins which are secreted by Vibrio cholerae and entertoxigenic Escherichia coli (ETEC), respectively.